Abstract
Introduction Acute myeloid leukemia (AML) is a fatal blood cancer with a 2-year survival rate of less than 50%. AML with TP53 mutations (TP53mut) has a particularly poor prognosis with a 1-year survival rate of less than 10%. Gene expression profiling (GEP) has been used to identify poor prognostic features in AML and other cancers. In acute lymphoblastic leukemia (ALL), GEP has been instrumental in defining new molecular subgroups with poor prognostic features such as Ph-like ALL. Because TP53mut AML is uniquely treatment refractory, we hypothesize that this AML subtype harbors a distinct GEP and TP53 wild type (TP53wt) AMLs that express features of the TP53mut GEP would have similarly poor clinical outcomes.
Methods and results: We analyzed RNA sequencing data of AML samples that have associated mutational profiling in the BEAT-AML (n=413) and TCGA (n=178) datasets. We compared the GEP of TP53mut and TP53 wild type (TP53wt) samples. Using unsupervised hierarchical clustering methods, we detected no significant clustering according to TP53 status. Therefore, we applied a machine-learning approach to detect whether a unique TP53mut GEP could be detected. Using the BEAT-AML dataset, we randomly divided the samples in half to generate training and testing datasets. The TCGA dataset was reserved as a validation dataset. We trained a Ridge regression model on the training dataset using 10-fold cross validation to define a model that could be used to classify TP53mut and TP53wt cases. We evaluated our model on the test dataset and found that this model was highly accurate in distinguishing TP53mut versus TP53wt cases (93% sensitivity and 97% specificity). When we applied this model to the validation dataset, we found a similar performance (93% sensitivity and 99% specificity) demonstrating that this model robustly defines a TP53mut GEP that generalizes across datasets.
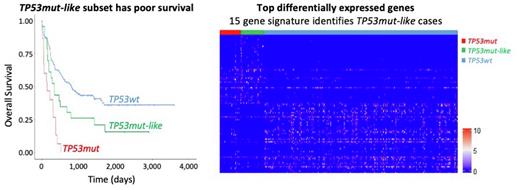
Although our machine learning model exhibited high accuracy in distinguishing TP53mut samples from TP53wt samples, we noted there was a cohort of TP53wt samples that were consistently scored highly by our classifier, suggesting they had features that made them TP53 mutant-like (TP53mut-like). Interestingly, we found that the TP53wt cases with the highest scores (representing highest similarity to the TP53mut profile) shared several clinical features with TP53mut samples. Most notably, the survival rate of TP53mut-like cases (TP53wt cases with high scores) was significantly lower than TP53wt cases with low scores. In the BEAT-AML dataset, we observed that the 42 TP53wt samples with the highest score had worse overall survival (OS) rates among the TP53wt cases (median survival: 347 days versus 568 days, p=0.014).
We reasoned that these TP53mut-like may represent a previously unappreciated subtype, and thus, we specifically trained a Ridge regression model to distinguish these TP53mut-like cases from TP53wt in the BEAT-AML dataset. This model was also highly accurate when tested on the held-out BEAT-AML samples (87.50% sensitivity and 95.52% specificity). When applying the resulting TP53mut-like model to the TCGA GEP data, we found that the 23 TP53wt patients with the highest scores had the worse OS rate among the TP53wt cases (median survival 335 days versus 800 days, p=0.031). Furthermore, we find that the TP53mut-like cases displayed other similar features to TP53mut cases. For example, TP53mut and TP53mut-like samples have higher LSC17 scores (Ng et al. Nature 2016) relative to TP53wt cases (p<0.0001). Clinically, we find that TP53mut cases have lower bone marrow blast percentage and white blood cell count (WBC) relative to TP53wt cases in both datasets (consistent with prior reports [Tashakori et al. Blood 2022]). Accordingly, TP53mut-like cases also have lower bone marrow blast percentage (35 versus 63%, p<0.0001) and WBC (22 versus 42%, p<0.0001) relative to TP53wt cases. The cytogenetic profiles of TP53mut-like cases resembled those of TP53mut cases, but these cytogenetic profiles were insufficient identify TP53mut-like cases and were detected in some TP53wt cases. Finally, we identified a panel of 15 genes whose expression identifies TP53mut-like cases.
Conclusion: We use a statistical regression model to define the GEP of TP53mut AML and define a new subset of TP53mut-like AML that lacks TP53 mutations but shares many of the same clinical and laboratory features of TP53mut AML, including poor survival.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal